16. XGBoost
XGBoost, or eXtreme Gradient Boosting, is gradient boosting library. Although scikit-learn has several boosting algorithms available, XGBoost’s implementations are parallelized and takes advantage of GPU computing. A few of the types of learners XGBoost has include gradient boosting for regression, classification and survival analysis (e.g. Accelerated Failure Time
AFT). There are no shortages of boosting libraries; here’s a few more.
16.1. Regression
Let’s see how XGBoost works on regression problems by first simulating data.
[1]:
import numpy as np
import random
from sklearn.datasets import make_regression
random.seed(37)
np.random.seed(37)
X, y = make_regression(**{
'n_samples': 1000,
'n_features': 10,
'n_targets': 1,
'bias': 5.3,
'random_state': 37
})
print(f'X shape = {X.shape}, y shape {y.shape}')
X shape = (1000, 10), y shape (1000,)
We will split the generated data into training and testing sets.
[2]:
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=37)
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
(800, 10) (800,)
(200, 10) (200,)
The XGBRegressor class is used to learn a boosted regression model. Note that the objective is specified to reg:squarederror. A list of objectives is available. Here, we specify only 10 estimators.
[3]:
import xgboost as xgb
model = xgb.XGBRegressor(objective='reg:squarederror', n_estimators=10, seed=37)
model.fit(X_train, y_train)
[3]:
XGBRegressor(base_score=0.5, booster='gbtree', colsample_bylevel=1,
colsample_bynode=1, colsample_bytree=1, gamma=0, gpu_id=-1,
importance_type='gain', interaction_constraints='',
learning_rate=0.300000012, max_delta_step=0, max_depth=6,
min_child_weight=1, missing=nan, monotone_constraints='()',
n_estimators=10, n_jobs=12, num_parallel_tree=1, random_state=37,
reg_alpha=0, reg_lambda=1, scale_pos_weight=1, seed=37,
subsample=1, tree_method='exact', validate_parameters=1,
verbosity=None)
We will measure the performance of the model using mean absolute error (MAE) and root mean squared error (RMSE).
[4]:
from sklearn.metrics import mean_squared_error, mean_absolute_error
y_pred = model.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
print(mae)
print(rmse)
35.00964208044854
44.165107049077356
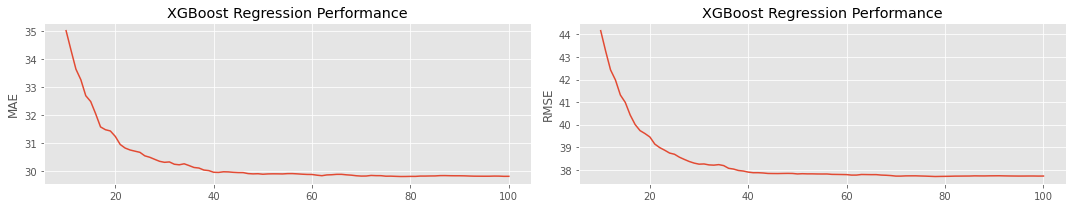
As you increase the number of estimators, the performance should increase, as measured by MAE and RMSE. There is a point of diminishing returns, however.
[5]:
import pandas as pd
def get_performance(n_estimators):
model = xgb.XGBRegressor(objective='reg:squarederror', n_estimators=n_estimators, seed=37)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
mae = mean_absolute_error(y_test, y_pred)
rmse = np.sqrt(mean_squared_error(y_test, y_pred))
return {'mae': mae, 'rmse': rmse}
n_estimators = list(range(10, 101, 1))
results = pd.DataFrame([get_performance(n) for n in n_estimators], index=n_estimators)
[6]:
import matplotlib.pyplot as plt
plt.style.use('ggplot')
fig, axes = plt.subplots(1, 2, figsize=(15, 3))
_ = results.mae.plot(ax=axes[0])
_ = results.rmse.plot(ax=axes[1])
axes[0].set_title('XGBoost Regression Performance')
axes[1].set_title('XGBoost Regression Performance')
axes[0].set_ylabel('MAE')
axes[1].set_ylabel('RMSE')
plt.tight_layout()
16.2. Classification
Let’s turn our attention to using XGBoost for classification by generating data for a classification problem.
[7]:
from sklearn.datasets import make_classification
X, y = make_classification(**{
'n_samples': 2000,
'n_features': 20,
'n_informative': 10,
'n_redundant': 0,
'n_repeated': 0,
'n_classes': 2,
'random_state': 37
})
print(f'X shape = {X.shape}, y shape {y.shape}')
X shape = (2000, 20), y shape (2000,)
We will split the data into training and testing sets.
[8]:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=37)
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
(1600, 20) (1600,)
(400, 20) (400,)
The input data must be transformed from numpy arrays into DMatrix.
[9]:
dtrain = xgb.DMatrix(X_train, y_train)
dtest = xgb.DMatrix(X_test, y_test)
print(dtrain.num_row(), dtrain.num_col())
print(dtest.num_row(), dtest.num_col())
1600 20
400 20
Now we are ready to learn a classification model with XGBoost.
[10]:
param = {
'max_depth':2,
'eta':1,
'objective':'binary:logistic',
'eval_metric': 'logloss',
'seed': 37
}
num_round = 20
model = xgb.train(param, dtrain, num_round)
We will measure the performance of the model using Area Under the Curve (the Receiver Operating Characteristic curve) and the Average Precision Score.
[11]:
from sklearn.metrics import roc_auc_score, average_precision_score
y_pred = model.predict(dtest)
auc = roc_auc_score(y_test, y_pred)
aps = average_precision_score(y_test, y_pred)
print(auc)
print(aps)
0.951115116017121
0.9450649144817633
16.3. Survival
Let’s see how survival regression works with XGBoost. Let’s sample some data.
[12]:
X, y = make_regression(**{
'n_samples': 1000,
'n_features': 4,
'n_targets': 0,
'random_state': 37
})
coef = np.array([2.0, -1.0, 3.5, 4.4])
baseline = np.e / np.power(1 + np.e, 2.0)
y = np.exp(-X.dot(coef)) * baseline
print(f'X shape = {X.shape}, y shape {y.shape}, coef shape = {coef.shape}')
X shape = (1000, 4), y shape (1000,), coef shape = (4,)
Here, we will split the data into training and testing sets.
[13]:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=37)
print(X_train.shape, y_train.shape)
print(X_test.shape, y_test.shape)
(800, 4) (800,)
(200, 4) (200,)
Let’s turn the inputs into the appropriate format. Note that we do not supply the duration into the DMatrix but set the lower and upper bound using the set_float_info() method.
[14]:
dtrain = xgb.DMatrix(X_train)
dtest = xgb.DMatrix(X_test)
dtrain.set_float_info('label_lower_bound', y_train)
dtest.set_float_info('label_lower_bound', y_test)
dtrain.set_float_info('label_upper_bound', y_train)
dtest.set_float_info('label_upper_bound', y_test)
print(dtrain.num_row(), dtrain.num_col())
print(dtest.num_row(), dtest.num_col())
800 4
200 4
The following shows how to specify a survival model with AFT.
[15]:
params = {
'objective': 'survival:aft',
'eval_metric': 'aft-nloglik',
'aft_loss_distribution': 'logistic',
'aft_loss_distribution_scale': 1.0,
'tree_method': 'exact',
'learning_rate': 0.05,
'max_depth': 5
}
model = xgb.train(params, dtrain, num_boost_round=40, evals=[(dtrain, 'train')])
[0] train-aft-nloglik:2.58474
[1] train-aft-nloglik:2.13443
[2] train-aft-nloglik:1.76805
[3] train-aft-nloglik:1.51551
[4] train-aft-nloglik:1.30791
[5] train-aft-nloglik:1.15456
[6] train-aft-nloglik:1.01593
[7] train-aft-nloglik:0.90408
[8] train-aft-nloglik:0.80288
[9] train-aft-nloglik:0.71937
[10] train-aft-nloglik:0.64163
[11] train-aft-nloglik:0.57327
[12] train-aft-nloglik:0.51536
[13] train-aft-nloglik:0.46241
[14] train-aft-nloglik:0.41428
[15] train-aft-nloglik:0.37151
[16] train-aft-nloglik:0.33085
[17] train-aft-nloglik:0.29569
[18] train-aft-nloglik:0.26403
[19] train-aft-nloglik:0.23559
[20] train-aft-nloglik:0.20816
[21] train-aft-nloglik:0.18294
[22] train-aft-nloglik:0.16056
[23] train-aft-nloglik:0.13968
[24] train-aft-nloglik:0.11968
[25] train-aft-nloglik:0.10160
[26] train-aft-nloglik:0.08428
[27] train-aft-nloglik:0.06922
[28] train-aft-nloglik:0.05431
[29] train-aft-nloglik:0.04075
[30] train-aft-nloglik:0.02782
[31] train-aft-nloglik:0.01519
[32] train-aft-nloglik:0.00409
[33] train-aft-nloglik:-0.00611
[34] train-aft-nloglik:-0.01670
[35] train-aft-nloglik:-0.02580
[36] train-aft-nloglik:-0.03482
[37] train-aft-nloglik:-0.04267
[38] train-aft-nloglik:-0.05033
[39] train-aft-nloglik:-0.05770
Below, we evaluate the model using the c-index.
[16]:
from itertools import combinations
def get_status(p1, p2):
x1, y1 = p1[0], p1[1]
x2, y2 = p2[0], p2[1]
r = (y2 - y1) * (x2 - x1)
if r > 0:
return 1
elif r < 0:
return -1
else:
return 0
y_pred = model.predict(dtest)
pairs = combinations([(y_t, y_p) for y_t, y_p in zip(y_test, y_pred)], 2)
results = [get_status(p1, p2) for p1, p2 in pairs]
c = sum([1 for r in results if r == 1])
n = len(results)
print(c / n)
0.9332160804020101
The c-index of the predictions from the training is as expected; it’s higher than the testing set.
[17]:
y_pred = model.predict(dtrain)
pairs = combinations([(y_t, y_p) for y_t, y_p in zip(y_train, y_pred)], 2)
results = [get_status(p1, p2) for p1, p2 in pairs]
c = sum([1 for r in results if r == 1])
n = len(results)
print(c / n)
0.9664893617021276